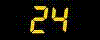1.基本概念
> clust.coeff<-function(matrix){
+ n<-nrow(matrix)
+ output<-rep(0,n)
+ for(i in 1:n){
+ m<-sum(matrix[i,])
+ output[i]<-
+ sum(t(matrix * matrix[i,])*matrix[i,])/(m*(m-1))
+ }
+ output[which(output=="NaN")]<-0
+ output
+ }
> A<-matrix(c(
+ 0,1,1,1,1,0,0,0,0,
+ 1,0,1,1,1,1,0,0,0,
+ 1,1,0,1,0,0,1,0,0,
+ 1,1,1,0,0,0,0,1,1,
+ 1,1,0,0,0,0,0,0,0,
+ 0,1,0,0,0,0,0,0,0,
+ 0,0,1,0,0,0,0,0,0,
+ 0,0,0,1,0,0,0,0,0,
+ 0,0,0,1,0,0,0,0,0),
+ nrow = 9)
> clust.coeff(A)
[1] 0.6666667 0.4000000 0.5000000 0.3000000 1.0000000 0.0000000 0.0000000
[8] 0.0000000 0.0000000
> mean(clust.coeff(A))
[1] 0.3185185
|
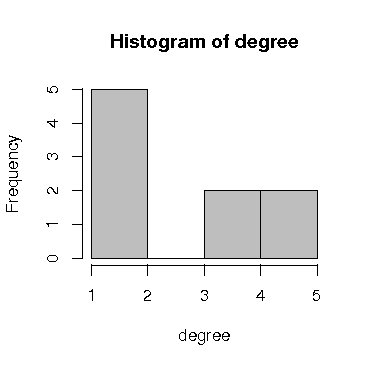
> library(sna) degree <- degree(A)/2 hist(degree, col = "grey") |
> rgraph(100,tprob=0.01,mode="graph")
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13]
[1,] 0 0 0 0 0 0 0 0 0 0 0 0 0
[2,] 0 0 1 0 0 0 0 0 0 0 0 0 0
[3,] 0 1 0 0 0 0 0 0 0 0 0 0 0
[4,] 0 0 0 0 0 0 0 0 0 0 0 0 0
[5,] 0 0 0 0 0 0 0 0 0 0 0 0 0
略
|
> erdos.renyi.game(100,p=0.01)
Vertices: 100
Edges: 51
Directed: FALSE
Edges:
[0] 4 -- 22
[1] 2 -- 23
[2] 6 -- 25
[3] 6 -- 27
[4] 0 -- 29
[5] 9 -- 30
[6] 9 -- 35
略
|
> random.graph.game(100, p = 0.01)
Vertices: 100
Edges: 35
Directed: FALSE
Edges:
[0] 1 -- 7
[1] 7 -- 8
[2] 15 -- 25
[3] 32 -- 40
[4] 19 -- 41
[5] 8 -- 44
|
> rg <- rgraph(100, tprob = 0.05, mode = "graph") > distance <- geodist(rg)$gdist > mean(distance[-c(which(distance == Inf), which(distance == 0))]) [1] 3.150484 > mean(clust.coeff(rg)) [1] 0.0345 > degree <- degree(rg)/2 > hist(degree, col = "grey") |
#レギュラーグラフ作成関数
regular.g <- function(n,k) {
reg <- matrix(0,n,n)
for (i in 1:n){
for (j in 1:k){
if (i+j <= n)
reg[i,i+j] <- 1
else
reg[i,i+j-n] <- 1
}
}
(reg <- symmetrize(reg))
}
#スモールワールド・シミュレーション
g <- regular.g(100,4)
Clustering <- 1:101
Distance <- 1:101
p <- seq(0,1,by = 0.01)
for (i in 1:101) {
clust <- 1:20
dist <- 1:20
for (j in 1:20){
rew.g <- rewire.ws(g, p[i])[1,,]
clust[j] <- mean(clust.coeff(rew.g))
d <- geodist(rew.g)$gdist
dist[j] <- mean(d[-c(which(d == Inf),which(d == 0))])
}
Clustering[i] <- mean(clust)
Distance[i] <- mean(dist)
}
x <- p; y <- Clustering
plot(x, y, xlab ="p", ylab = "Clustering Coefficient")
x <- p; y <- Distance
plot(x, y, xlab ="p", ylab = "Average Distance")
x <- p
y <- matrix(c(Clustering/Clustering[1],Distance/Distance[1]),101,2)
matplot(x, y, log = "x", pch = c(1,2), xlab ="p", ylab = "", col = "black")
legend(locator(1), pch = c(1,2),
c("clustering coefficient","average distance"))
|
g <- barabasi.game(10000, m = 2, directed = FALSE) dd <- degree.distribution(g) plot(dd,log ="x", xlab = "degree", ylab = "proportion") plot(dd,log ="xy", xlab = "degree", ylab = "proportion") average.path.length(g) clust <- transitivity(g,type ="local") clust[which(clust == "NaN")] <- 0 mean(clust) |
可視化にCytoscapeを利用。 この例では10,000ノードのスケールフリーネットワークの作成し、 ネットワークをテキストデータとしてエクスポート。
> library("igraph")
> graph2<-barabasi.game(1000,power=1)
> write.graph(graph2,"table2.txt","edgelist")
|
CytoscapeでFile-->Import-->Network from Tableよりインポート。

Back to R